bringing data and tools together
Genboree is a web-based platform for multi-omic research and data analysis using the latest bioinformatics tools.
You can upload your data and perform various analyses using a “drag and drop” user interface. Keep it private or share with collaborators. Bioinformatics tools and computational infrastructure are available for researchers who may not have programming expertise, or the time to pursue technical programming and/or scripting.
Get Startedworkbench
BIOINFORMATICS TOOLS
FOR BIOMEDICAL
DISCOVERY
The Genboree Workbench manages the many technical software and hardware aspects of genome-centric research for end-users. The Workbench contains bioinformatics tools useful for analyses in genomics, epigenomics, metagenomics, and transcriptomics, and a “drag and drop” interface makes the Workbench easy to use.
The Epigenome Toolset
Tools for analyzing DNA methylation and histone marks at the level of whole genomes, pathways, gene elements (i.e.. promoters, enhancers), and genetic loci to identify biological patterns. Read more about the Epigenomics Toolset
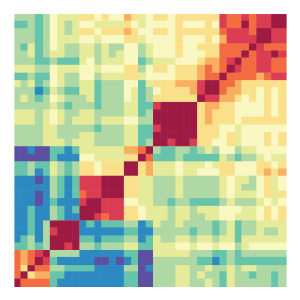
Heatmap:
The heatmap tool performs hierarchical clustering based on pairwise correlation of normalized epigenomic data sets. Users can choose from many available methods implemented in R for normalization, correlation, distance measure calculation, hierarchical clustering, and visualization.
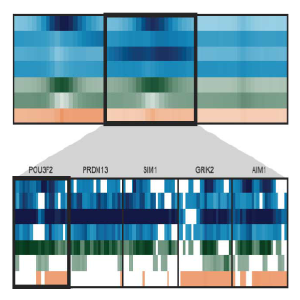
Spark:
Interactive visualization for k-means clustering of epigenomic data. Spark helps discover and visualize patterns of epigenomic profiles on a genome-wide scale. Regions of interest (i.e. promoters, enhancers, etc) can be downloaded in the form of annotation tracks for downstream analyses.
The Transcriptome Toolset
Tools and pipelines for RNA-Seq data analysis. More details coming soon...
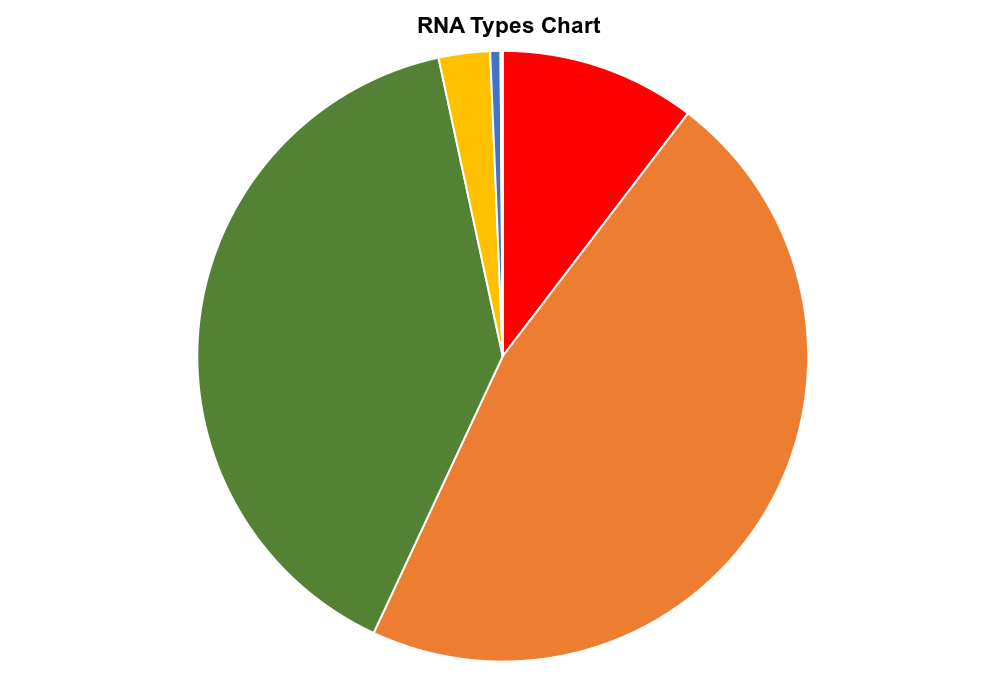
Small RNA-seq pipeline:
The small RNA-seq pipeline is for the processing and analysis of RNA-seq data generated to profile small exRNAs. It can handle multiple libraries and outputs abundance estimates, a variety of quality control metrics such as read-length distribution, summaries of reads mapped, and detailed information for each read mapped to each library.
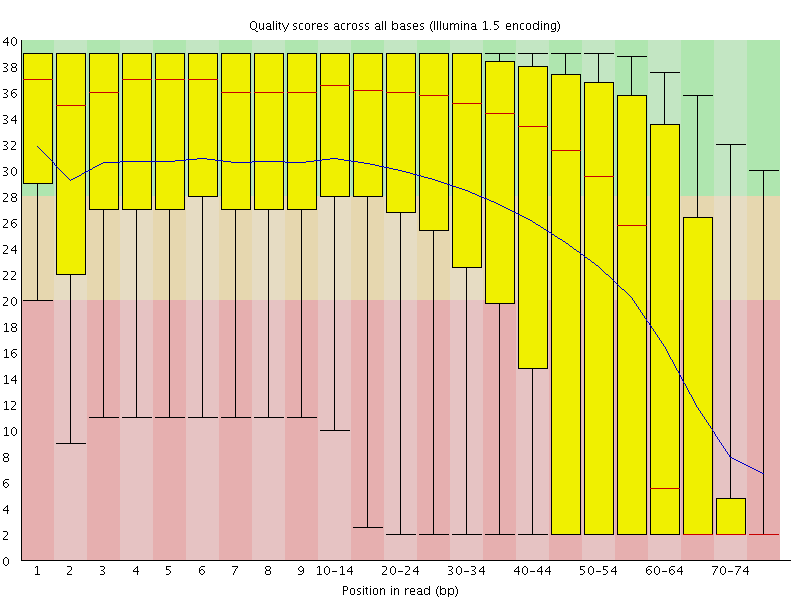
Long RNA-seq pipeline:
The long RNA-seq pipeline is for the processing and analysis of RNA-seq data generated from long-RNAs. The pipeline performs a quality check (FastQC), maps reads to a reference genome (Bowtie2), and post-processes the aligned reads (Samtools). The pipeline also performs gene-expression quantification, generates tracks for visualization, calculates mapping bias, and computes annotation coverage (all using RSEQtools).
The Microbiome Toolset
An interactive environment to conduct 16S rRNA microbiome and whole genome shotgun (WGS) sequencing analyses. The Toolset drives hypothesis generation by providing a wide range of analyses for studying metagenomic datasets. More details coming soon...

16S rRNA:
The 16S rRNA metagenomics toolset equips a researcher to explore their microbiome samples for a variety of analysis types such as alpha diversity, beta diversity, phylogenetic profiling, supervised machine learning, and feature selection.
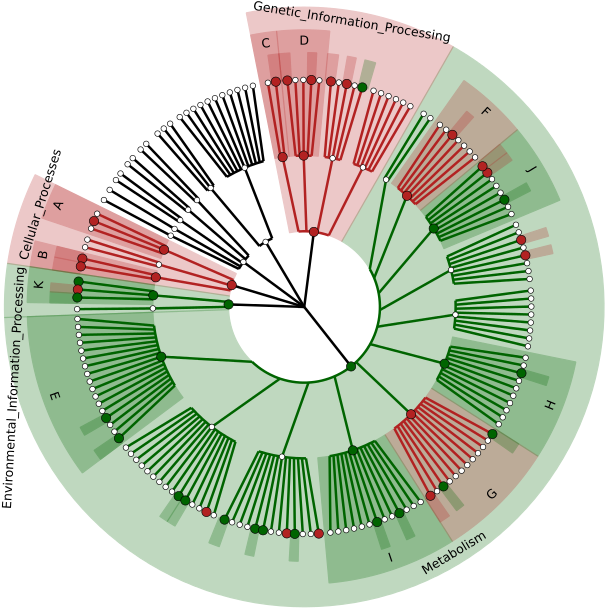
Shotgun Metagenomic Sequencing (WGS) (Coming Soon):
The shotgun metagenomic sequencing toolset empowers users to explore ‘what is there’ and ‘what are they doing’ types of questions. Taxonomic identity assesses identifiable organisms and results in summary matrices (relative percentages), biomarker discovery, heatmaps, and cladograms. Functional annotation identifies potentially active pathways from KEGG orthologous genes in which to compare and contrast sample groups.
commons
Online collaboration
The Genboree Commons is a place to create projects, discussion forums and wikis, and to share documents with your colleagues.
All of your documents and content are private within a project unless you choose to share with others. It’s easy to add colleagues to your projects, change access privileges, and communicate with one another.
ClinGen
Clinical Genome Resource
One of the main goals of the Clinical Genome Resource project is to enable curators and researchers to understand the clinical significance of genes, variants and genetic disorders. We contribute to this effort by developing several software products, web services and user interfaces to facilitate curation of genes and variants.
Learn more about our software products developed for ClinGen
THE HUMAN
EPIGENOME ATLAS
A Data Repository of Tissue-specific Epigenomic States
The Human Epigenome Atlas contains human reference epigenomes and the results of integrative and comparative analyses. Atlas data provides detailed insights into locus-specific epigenomic states, including histone marks and DNA methylation across tissues, cell types, developmental stages, physiological conditions, genotypes, and disease states.
Research
 Image: National Institutes of Health
Image: National Institutes of Health
NIH ExRNA Communication Program
The goal of the ERCP is to better understand the fundamental biological mechanisms of extracellular RNA (exRNA) generation, secretion, and transport, to create a public dataset of where exRNAs exist in normal human body fluids, and to explore their potential as therapeutics and biomarkers.
 Image: National Institutes of Health
Image: National Institutes of Health
The NIH ClinGen Resource
One of the components of the Clinical Genome Resource project is ClinGenDB infrastructure to enable the development of a knowledge base about genetic variants of clinical significance. ClinGenDB infrastructure consists of databases and web services implemented using Genboree KnowledgeBase (GenboreeKB).
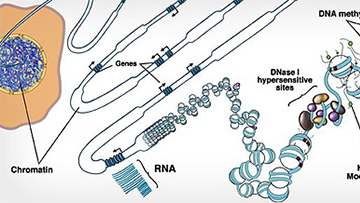 Image: National Institutes of Health
Image: National Institutes of Health
NIH Roadmap Epigenomics Mapping Consortium
The goal of this consortium is to map DNA methylation, histone modifications, chromatin accessibility, and small RNA transcripts in tissues and organs frequently involved in human disease.